Deciphering shell proteome within different Baltic populations of mytilid mussels illustrates important local variability and potential consequences in the context of changing marine conditions
Deciphering shell proteome within different Baltic populations of mytilid mussels illustrates important local variability and potential consequences in the context of changing marine conditions
Deciphering shell proteome within different Baltic populations of mytilid mussels illustrates important local variability and potential consequences in the context of changing marine conditions. J. Aviralagan, B. Marie, G. Chiapetta, J. Vinh, X. Gallet, M. Lebon, S. M'Zoudi, P. Dubois, S. Berland, A. Marie. Science of the Total Environment 745 (2020). doi.org/10.1016/j.scitotenv.2020.140878
Bivalve molluscs defend themselves against predation and environmental stressors through a mineralized shell. The noteworthy success of the Mollusca through time and space is claimed to be substantially due to their shells. Yet, the calcified shells are now seen as their potential vulnerability under environmental change with unbalanced ions and the decreasing calcium carbonate saturation state in seawater. Skeletons of marine species may become burdensome, energetically expensive to produce and less sharpened due to challenging their maintain conditions. There is an increasing concern about their fate and mussels are widely used in global change effects follow-up. Shell matrix proteins are part of the molecular control regulating the biomineralization processes for shell production and their differential expression may lead to the phenotypic plasticity of shells.
Here we examine how the modulation of the shell matrix proteins relates to shell formation by comparing North Sea mussels living in complete marine environment with Baltic Sea mussels that are subjected to waters with high riverine input and lower buffering capacity and how it alters the physical and biochemical properties of the shells.
The outcome was more fragile shells in mussels inhabiting Baltic Sea. Modulation of proteins comprising the biomineralization tool kit is observed. These data showed a relative increase in chitin related proteins, decrease in SD-rich, GA-rich proteins indicating altered protein scaffolding and mineral nucleation which lead to impaired shell microstructures influencing shell resistance in Baltic mussels. Interesting fact : shell matrix proteins with immunity domains are also found to be modulated.
Shell traits such as periostracum thickness, organic content and fracture resistance qualitatively correlates with the modulation of SMPs providing key insights into control of biomineralization at molecular level in the context of changing marine conditions.
Legend: Baltic mussels illustrates the shell proteome response to the environment and the potential consequences in the context of changing marine conditions.
Relationship between shell properties and shell proteome in Baltic Sea blue mussels living in natural brackish water in comparison with North Sea mussels living in complete marine environment is investigated in the context of changing ocean. Modulation of shell matrix proteins influence the mechanical properties and phenotype traits of the shells. Biochemical defense offered by the shell matrix proteins are balanced with respect to salinity and habitat.
Picture gallery :
Picture 1 : Scanning electron microscopy images show altered microstructure of Baltic mussel shells. The region of the shell used for acquiring the images is the interface of the nacreous layer, myostracum and prismatic layer of the shells from the North sea and the Baltic sea (Nynäshamn site). In Baltic mussels the nacreous layer is formed by relatively thin aragonite tablets compared to North Sea mussels (arrows). © Sophie Berland
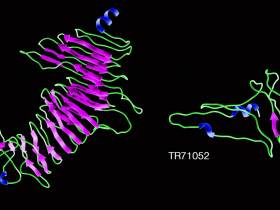
Picture 2 : Predicated structure of scaffolding shell matrix proteins TR78416 and TR71052 predicted using the I-Tasser tool. TR78416 proteins contain motifs that are formed by β-strand-turn-β-strand and are similar to that of spider silk. Primary protein sequences present low complexity domains with the GGXGG repeating motifs. © Sophie Berland
BOREA contact: Sophie Berland